Timothy (arguing in defense of the 'Intelligent Design' hypothesis): "Evolution has certain severe problems to overcome before I am convinced:
1. Mutations -- Mutations are the only way to introduce new genetic material into an organism. Thus, mutations are the driving force of Evolutionary theory. The problem with mutations are that the vast majority -- like a thousand to one -- are harmful. Thus, mutation is far more likely to destroy a species than cause it to evolve. If you throw a wrench at a car, you my fix it, but you are much more likely to damage it than fix it. To believe that a proven destructive force like mutation will eventually build a single complex living organism, much less ALL living organisms everywhere, is an act of faith, not of science."
Sharon: I happen to know somebody who has a mutation that is "harmless". I do not know where Timothy gets his figures from about "a thousand to one" are harmless.The conversation called to mind what John F. Kennedy said about our relationship to the sea:
"I really don't know why it is that all of us are so committed to the sea, except I think it's because in addition to the fact that the sea changes, and the light changes, and ships change, it's because we all came from the sea. And it is an interesting biological fact that all of us have, in our veins the exact same percentage of salt in our blood that exists in the ocean, and, therefore, we have salt in our blood, in our sweat, in our tears. We are tied to the ocean. And when we go back to the sea -- whether it is to sail or to watch it -- we are going back from whence we came." --John F. Kennedy, Jr., 1962[Though the scientific data might have changed since 1962, the fact of our relationship with the sea, has not.]
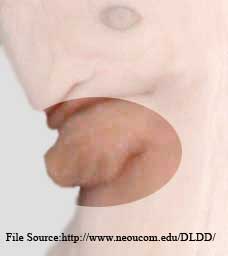
Of course the photographs we've collected of hind limbs on whales, and recently the flippers with toe nail photos on sea cows.
I called the woman I know and we photographed her and her daughter's webbed toes. My question is sensible enough. If two types of mammals (Sirenians and Cetaceans) could return to the sea, and evolve webbing between their fingers or toes (eventually turning into a tail or flippers), couldn't this mutation show up in humans as well? Sirenians, Cetaceans and Humans are all mammals... it would seem they could have the same genetic mutations occur? I think these "webbed toes" are genetically, even if just remotely perhaps, some insight into land to sea mammal evolution. I also have a photograph of a dolphin embryo where the fingers are evident and they appear to be webbed [obtained from J.G.M. Thewissen's Digital Library of Dolphin Development] -- as you guys know even the flippers of whales have bones in them for digits (their evolutionary past with land mammals).
Is there a relation between human webbed toes and the webbing that occured to transitional land to sea mammals?
Thanks, Sharon
I happened to have known somebody who has a mutation that is "harmless". I do not know where Timothy gets his figures from about "a thousand to one mutations" are harmless.
Of course you know someone with a harmless mutation. Everybody has many many harmless mutations. Hair color, eye color, skin tone etc. are all mutations that are essentially harmless.
The fins of Cetaceans don't have the digits spread out but rather a fairly tight mass with the digits all together. I'm not as familiar with Piniped morphology.
The major changes to adapt to a marine lifestyle seem to be streamlining of the body, heat retention adaptations and metabolic and lung changes to allow more time between breaths. While aquatic birds and otters do have webbed digits I'm not sure if there is any evidence for it being a major factor in cetacean evolution.
-Ken Shaw (talk.origins)
|
Website on "Syndactyly" My web site is a very modest effort to help people with syndactyly, and mothers of babies with syndactyly understand the condition better. Sort of an informal, unorganized self-help group. Recently, I received some input from a couple of ladies in India, one a Muslim college student. One does the correspondence for both. This helps to show how this is a global phenomenon which can and does happen "anywhere". I am thinking about changing part of the site to call itself the "World-Wide-Webbed-Toes" site. Your page seems to be directed to explaining syndactyly to people who do not have it, which is great. The people who visit my pages will still benefit from the pictures you provide. The more the better. One of the most common questions (Syndactyly FAQ?) concerns whether this is inherited. I tell people "yes and no". In some cases, there are too many close relatives with this condition to be explained by chance. Other cases, like mine, seem to be isolated, with no known relatives showing this condition. Neither of my children have syndactyly. I am still waiting for grandchildren. We will see what turns up when that happens. Best regards, Bill Luken |
Is there a relation between human webbed toes and the webbing that occured to transitional land to sea mammals?
I believe there is. Developmentally it's just the turning off of apoptosis between the digits during early development. There are doubtless many different mutations, at many different parts of the developmental sequence, that could accomplish that same trick. Junk DNA would have nothing to do with it, and it's unlikely that the same mutation would be involved since there are probably so many possible ones.
But the biggest problem with the creationist spiel has nothing to do with the frequency of neutral, beneficial, or deleterious mutations. (And remember that these categories are dependent on environment, by the way.) The biggest problem is a simple failure to understand natural selection. So what if the ratio of good to bad is 1:1000 (or whatever ratio you like)? Natural selection gets rid of the bad ones when they're at low frequencies, and promotes the good ones to high frequencies. Thus a mutation won't destroy a species; it may destroy an individual, but that's how natural selection works. The species as a whole collects good mutations, however rare they may be, through the differential reproductive success of individuals. How can one possibly criticize evolution without understanding its most basic principles?
-John Harshman (talk.origins)I wonder if the same developmental mutation in your friend's gene happened in the homologous region of the dolphin's DNA? It would take a lot of research to discover that of course.
We share lots of genes with mice, probably many with cetaceans too, since they were originally mammals and all mammals share a common ancestor. Some of those genes probably direct whether the skin between the finger bones dies or not. I was just wondering what the difference was between the genes that directed the development (and disappearance) of the skin between the fingers in human beings and cetacea. It's not something I know anything about, and I haven't heard that any cetacean genomes have been sequenced yet. Though they did sequence the human genome, the whole thing, and chimp genome, and rat genome, and mouse genome and chicken genome (very recently, this month in fact) and zebra-fish genome (ancient cousin of the fish that led to the first amphibians). I don't know what others are due next for sequencing. But the results so far don't pose any problems for evolutionists, since a lot of the same genes or very similar ones, have been found, even in creatures as far apart as mice and men. Recently a few developmental genes of the only known living species of lobe-finned fish have been sequenced. Not the whole genome, just a few genes, crucial developmental ones that play a role in fore-limb formation, because the lobe-finned fish preceded the earliest amphibians in the fossil record. And, as expected, the genes for the development of the forelimb of lobe-finned fishes more closely resembled those of mice than of other fishes (with their non-lobed-fore-fins). I am still collecting articles on all of this stuff. Oh, the guy who headed up the human genome sequencing project is a Christian, a theistic evolutionist. He thinks the creationists are an embarrassment to his faith.
-Ed BabinskiEmail to Professor Thewissen
December 19, 2004
Dolphin Embryo Picture (Permission to use?)Dear Professor Thewissen,
I had a conversation with a fellow who believes in Intelligent Design and part of his argument against Darwinism was "mutations are harmful". I got to thinking about a person I personally know who had a genetic mutation she was born with on her feet, and she passed this gene on to her daughter. They both have webbed toes, which appears similar to the process that took place on cetaceans and sirenians, with the digits webbing together to form a flipper... so I got photographs and created a page, and collected some comments from talk.origins on the question. (At least I feel there is something in common here... I would assume since cetaceans, sirenians and humans are all mammals, the same phenomena of feet to flippers, could [at least, hypothetically] happen.)
I have borrowed the picture of the dolphin embryo with the webbed fingers, from your site (and placed proper credit to your URL on Dolphin Embryo Development site). I hope this is okay. By the way, you guys have done a wonderful job on that site.
I hope there is no problem that I borrowed that photograph. I wanted to drop a line, just in case there was any problem about it. I've read on other pages from your site, that the photographs were accessible, as long as proper credit is given. Please let me know if there is any problem.
Sincere appreciation,
Sharon Mooney"J. G. M. Thewissen"
Tuesday, December 21, 2004
Subject: Re: Dolphin Embryo Picture (Permission to use?)No great, please use that image on your page.
You have to be careful. A number of genes are involved, and the mechanism whereby a dolphin retains webbed feet might be very different from the mechanism whereby some humans retain webbed feet. For the dolphin, we don't know which genes are involved.
Are you aware that all mammal embryos have webbed feet early in development, and then the skin flaps between the fingers die.
Just making sure.
Hans Thewissen"ed.babinski/furman.edu"
Tuesday, December 21, 2004
Subject: Re: Fw: Dolphin Embryo Picture (Permission to use?)He's right. All embryos have skin between the finger bones and that skin undergos cell death and shrivels away. I would assume it's the same genes or very similar genes in all mammals that direct the cell death process that makes the webbed skin die between the finger bones. If that gene gets mutated (or another gene related to that process) then the webbed skin would remain.
The whale's skeleton closely resembles the skeletons of other mammals. For instance, the bones of the flippers resemble jointed limbs and digits and the neck has seven vertebrae like many other mammal species including man.
(Even the giraffe's neck has the same standard number of vertebrae, seven!)[See The Macmillan Illustrated Encyclopedia of Dinosaurs and Prehistoric Animals: A Visual Who's Who of Prehistoric Life, Collier Books: New York, 1988, pg. 230-231]
In mammalian embryos, the forelimb initially forms as a small bump on the side of the chest, the so-called limb bud. The limb bud grows and flattens, and fingers become apparent. At later stages in dolphins (third and fourth specimen above) the fingers can be seen as individual entities, as they are in most mammals. Lateron, they disappear and are all embedded in a narrow, fin-like extremity called the flipper. If evolution is true and these tiny fingers are a glance back into the evolutionary history of dolphins, could it happen to humans? Below are photographs of the genetic mutation which occured when a non-webbed toed mother gave birth to a daughter who had the webbed toe gene, and twenty five years later passed that gene on to her daughter. |
|||
 |
|||
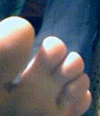
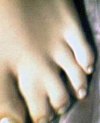
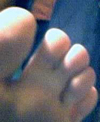
Photograph of mother and daughter. The Grandmother did not have webbed toes, and none of her relatives or immediate ancestors were known to have webbed toes. For both women, the genetic mutation is located on both feet, affecting the same toes. |
|||
|
|||
 |
|||
Daughter's webbed toe. This webbing occurs on both feet, in the same location. |
|||
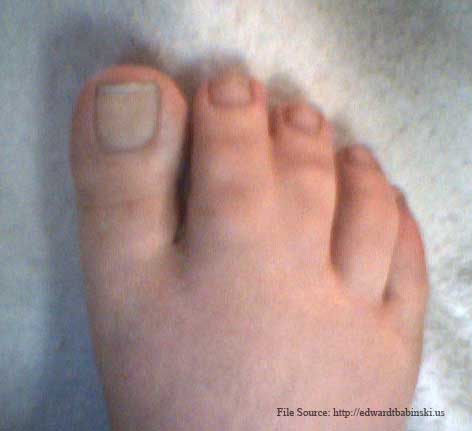 |
|||
Mother's webbed toe. This webbing occurs on both feet, in the same location. |
|||
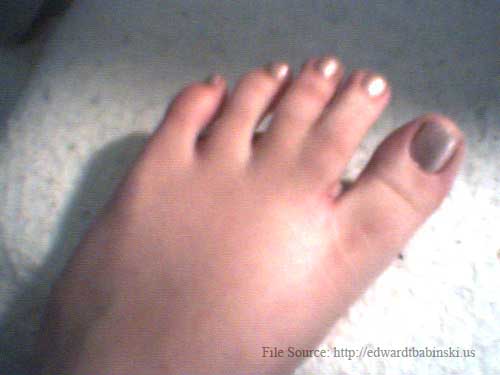 |
|||
Daughter's webbed toe, on left foot. |
|||
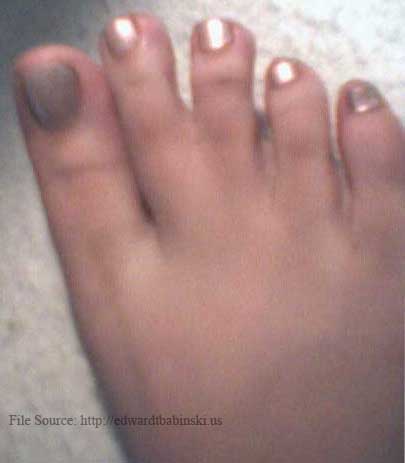 |
|||
Daughter's webbed toe, on right foot. |
 |
Edward B.: I don't know if humans could evolve into cetaceans. Maybe, maybe not. There's limitations to consider. Usually species don't go back and re-evolve into one another. We share a common mammalian ancestor with cetaceans in the past, but species usually continue to diverge and go off in different directions. Sharon M.: Ed, I may not have a degree in biology but I'm not stupid. Of course humans wouldn't evolve into Cetaceans or Sirenians. What would they call a human with webbed hands and feet / fluke. Ah, yes, a mermaid ! Edward B.: Dugong you're right! |
"My Webbed Toes Are Cool"
February 22, 2005, "Joe" wrote:
Thems not webbed feet its just two deformed toes that grew togeather, webbed toes have a thin skin in between those toes are joined togeather, big differance.Hi Joe, besides misspelling, I feel you have no real understanding of what syndactyly is. But thanks for the input.
You need to refer to Dictionary com if you have any further questions and go from there.
Syndactyly is not a simple "deformation". In fact, individuals with syndactyly would disagree with you about your callous label of their condition as "deformed". The lady in the pics above expressed having always felt her webbed toes were "neat" (in a cool kind of way). It is genetic and can be passed generation to generation. *smile*.
1. Hereditary and Genetic
2. Referred to as "Webbing" of Digits
3. It is not an isolated deformation and the proper scientific name for the condition is "syndactyly."
4. Found in mammals and birds
syn·dac·ty·ly ( P ) Pronunciation Key (sn-dkt-l) or syn·dac·tyl·ism (-t-lzm) n. Biology
The condition of having two or more fused digits, as occurs normally in certain mammals and birds.
A congenital anomaly in humans characterized by two or more fused fingers or toes.Main Entry: syn·dac·ty·ly
Pronunciation: -lE
Function: noun
Inflected Form: plural -lies
: a union of two or more digits that is normal in many birds (as kingfishers) and in some lower mammals (as the kangaroos) and that occurs in humans often as a hereditary disorder marked by the joining or webbing of two or more fingers or toessyndactyly
n : birth defect in which there is partial or total webbing connecting two or more fingers or toes [syn: syndactylism]syn·dac·ty·ly (sn-dkt-l)
n.
Webbing or fusion of the fingers or toes, involving soft parts only or including bone structure. Also called symphalangism, syndactylia, syndactylism, zygodactyly.
Source: dictionary.reference.com